Instances
Models
Five well known biological models:
- FY : Fission Yeast by Davidich and Bornholdt (2008) (doi:10.1371/journal.pone.0001672)
- SP : Segment Polarity (1 cell) by Sánchez et al. (2002) (doi:10.1387/ijdb.072439ls)
- TCR : TCR Signalisation by Klamt et al. (2006) (doi:10.1186/1471-2105-7-56)
- MCC : Mammalian Cell Cycle by Fauré et al. (2006) (doi:10.1093/bioinformatics/btl210)
- Th : T-helper Cell Differentiation by Mendoza and Xenarios (2006) (doi:10.1186/1742-4682-3-13)
Models were corrupted according to different parameters:
- F : probability of changing a regulatory function
- E : probability of flipping the sign of an edge
- R : probability of removing a regulator
- A : probability of adding a regulator
24 different configurations of corruption parameters were considered and 100 model instances were generated for each configuration.
Generated models can be found here.
Observations
Five sets of observations were generated consistent with the original models under the following update schemes:
- Synchronous
- Asynchronous
The observations were generated with a length of 20 time-steps. For the experimental evaluation it were considered also a part of the same observations with only 3 time-steps.
Generated observations can be found here.
Results
For detailed time results, please check results-global-v1.3.1.pdf.
obs - number of observations
ts - number of time-steps
Synchronous
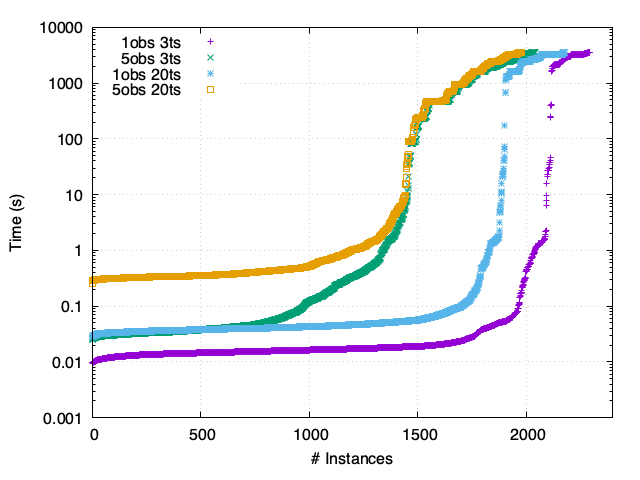 FY - Synchronous
FY - Synchronous
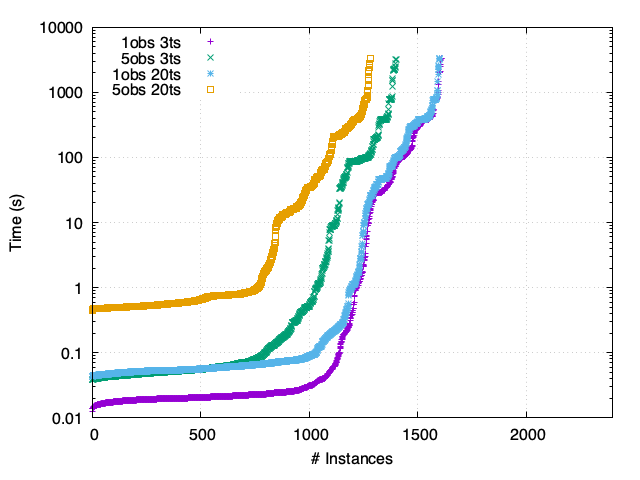 SP - Synchronous
SP - Synchronous
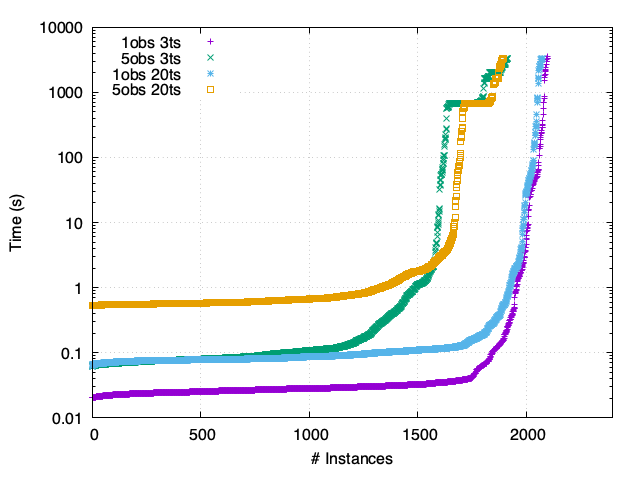 TCR - Synchronous
TCR - Synchronous
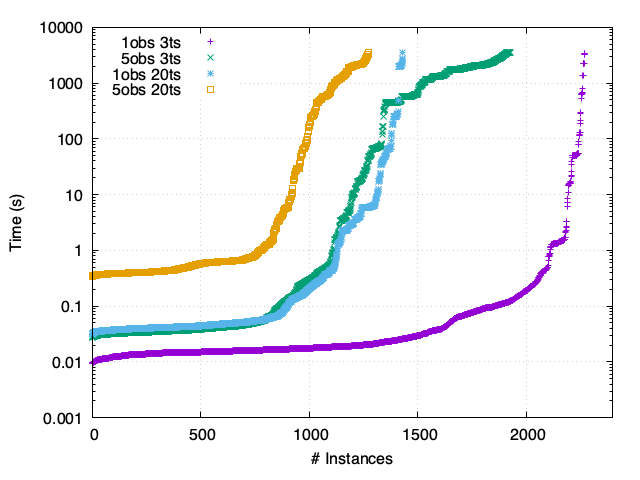 MCC - Synchronous
MCC - Synchronous
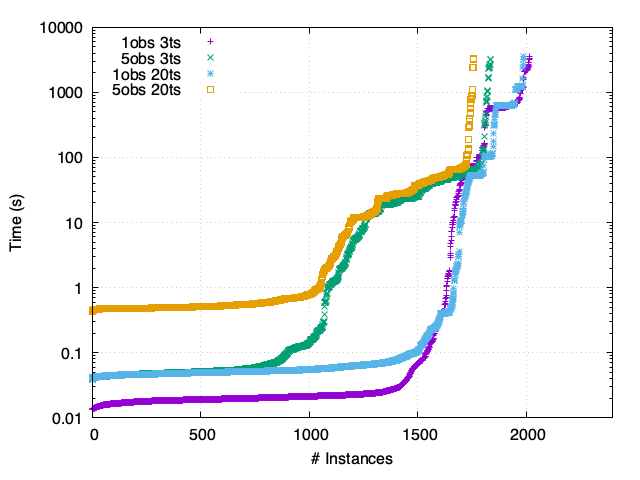 Th - Synchronous
Th - Synchronous
Asynchronous
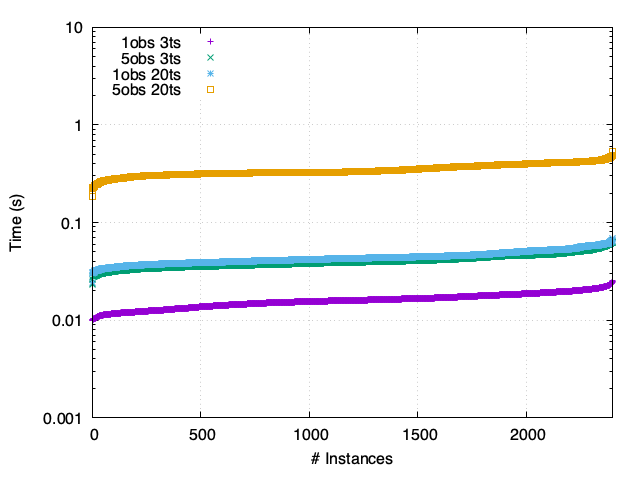 FY - Asynchronous
FY - Asynchronous
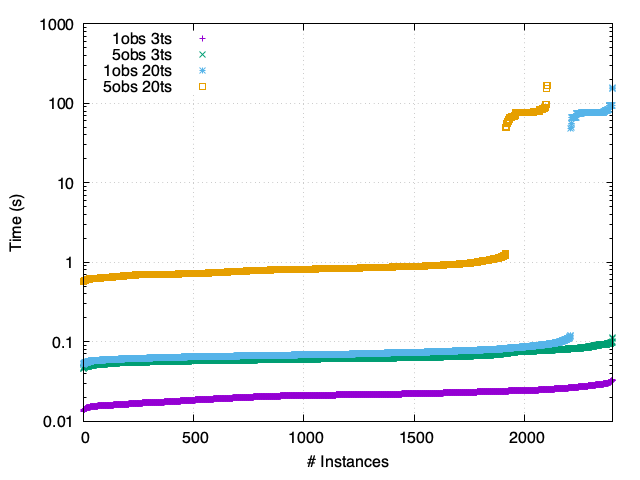 SP - Asynchronous
SP - Asynchronous
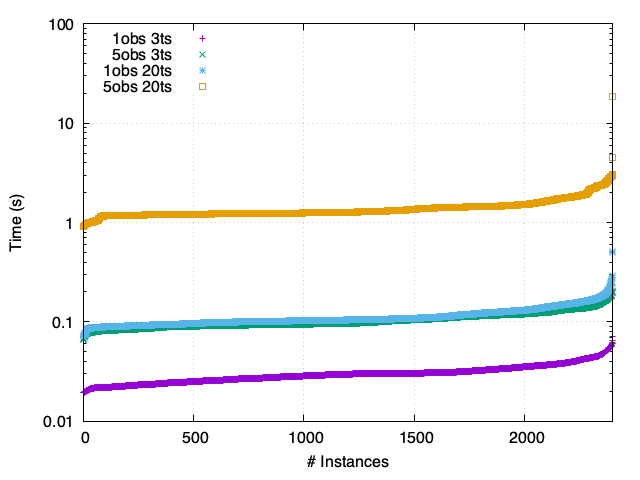 TCR - Asynchronous
TCR - Asynchronous
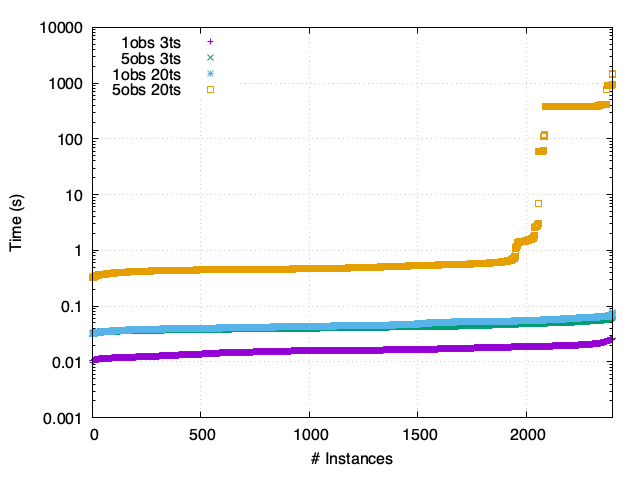 MCC - Asynchronous
MCC - Asynchronous
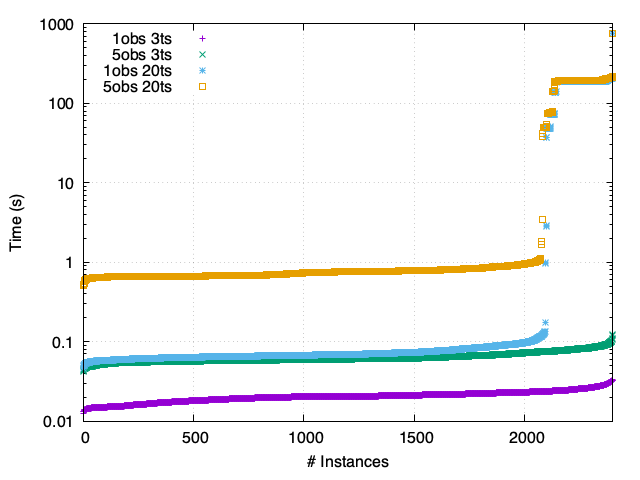 Th - Asynchronous
Th - Asynchronous